This post was imported from my old Drupal blog. To see the full thing, including comments, it's best to visit the Internet Archive.
My previous post talked about how to install 4store as a triplestore, and use the Ruby library RDF.rb in order to process RDF extracted from that store. This was a response to Richard Pope’s Linked Data/RDF/SPARQL Documentation Challenge which asks for documentation of how to install a triplestore, load data into it, retrieve it using SPARQL and access the results as native structures using Ruby, Python or PHP.
I quite enjoyed writing the last one, so I thought I’d try again. As before, I am on Mac OS X, but this time I’m going to use Python, which I have not programmed in before. I like a challenge. You might not like the results!
Sesame
This time, I’m going to use Sesame, as I was told by John Sheridan that it was so easy to install that even he, a civil servant, could do it!
Sesame needs a Java servlet container. I’m using Tomcat because I have some experience with it, but you could use something like Jetty if you prefer. I had a bit of trouble getting Tomcat 6 to install, but that might just have been because it has a lot of dependencies and I wasn’t patient enough. It might be worth upgrading your existing ports and getting verbose output so you know there’s activity as you install Tomcat:
$ sudo port upgrade outdated
$ sudo port -v install tomcat6
This installs Tomcat 6 in /opt/local/share/java/tomcat6.
While that’s happening, get Sesame from its download page. I got hold of openrdf-sesame-2.3.2-sdk.tar.gz. The files we actually need are the .wars so we can just extract them and put them in the webapps directory within Tomcat:
$ tar -zxvf openrdf-sesame-2.3.2-sdk.tar.gz openrdf-sesame-2.3.2/war/*.war
$ sudo cp openrdf-sesame-2.3.2/war/*.war /opt/local/share/java/tomcat6/webapps/
Then startup Tomcat:
$ sudo /opt/local/share/java/tomcat6/bin/tomcatctl start
All being well, you should see Tomcat doing some initial setup:
conf_setup.sh: file conf/catalina.policy is missing; copying conf/catalina.policy.sample to its place.
conf_setup.sh: file conf/catalina.properties is missing; copying conf/catalina.properties.sample to its place.
conf_setup.sh: file conf/server.xml is missing; copying conf/server.xml.sample to its place.
conf_setup.sh: file conf/tomcat-users.xml is missing; copying conf/tomcat-users.xml.sample to its place.
conf_setup.sh: file conf/web.xml is missing; copying conf/web.xml.sample to its place.
conf_setup.sh: file conf/setenv.local is missing; copying conf/setenv.local.sample to its place.
Starting Tomcat.... started. (pid 20064)
Now have a look at http://localhost:8080/openrdf-sesame. If you’re like me, you’ll get some error messages because the user that Tomcat is running under (www) isn’t able to create or write to a logging directory that it wants to create, in my case /Users/Jeni/Library/Application Support/Aduna/OpenRDF Sesame/logs. This turns out to be partly caused by permissions issues and partly caused by the spaces in the filename. To get around it, create a data directory for Aduna that doesn’t have spaces in the filename, and change its ownership to www. In my case, I chose /opt/local/var/aduna.
$ sudo mkdir -p /opt/local/var/aduna
$ sudo chown www:www /opt/local/var/aduna
Then edit Tomcat’s setenv.local file which in my environment is at /opt/local/share/java/tomcat6/conf and add a line that sets the info.aduna.platform.appdata.basedir to that directory, like this:
export JAVA_OPTS='-Dinfo.aduna.platform.appdata.basedir=/opt/local/var/aduna'
Restart Tomcat:
$ sudo /opt/local/share/java/tomcat6/bin/tomcatctl restart
Then navigate again to http://localhost:8080/openrdf-sesame and you should see the Welcome page:
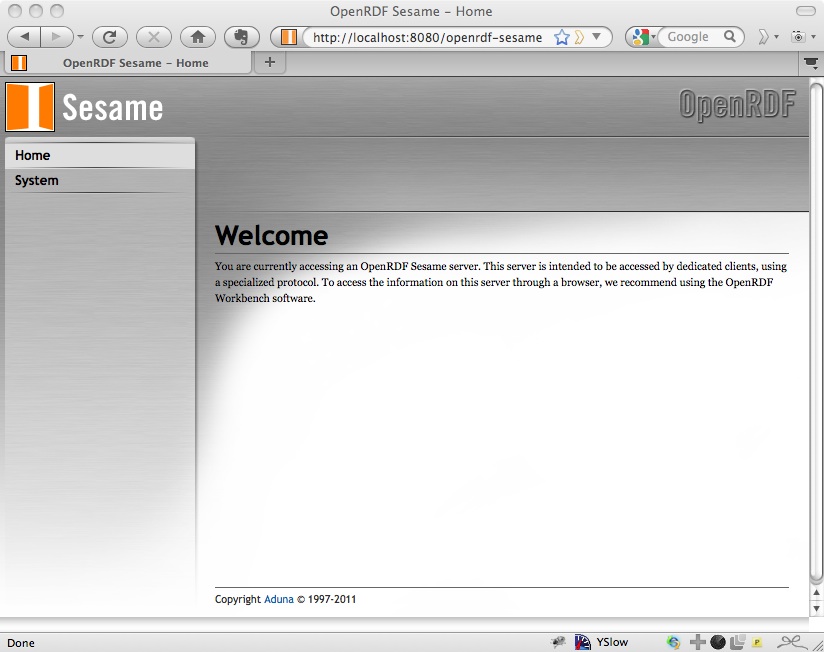
As you can see, this recommends using the Workbench for managing the repositories. If you open that, at http://localhost:8080/openrdf-workbench.
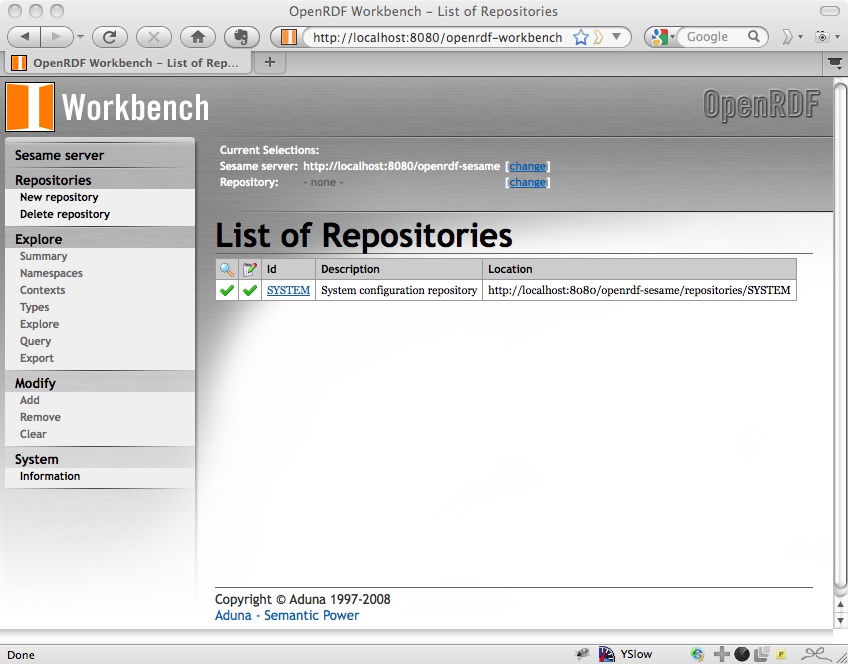
We’ll use this Workbench to create a new repository for our data, which I’ll call reference. Click on New Repository from the left hand navigation and fill in the form. I’m just going to use the default in-memory RDF store because I’m only using a little data; the other options (using MySQL or PostgreSQL stores) would be useful if I were creating something more permanent. See the Sesame User Guide for information about those.
So fill in the form to create a new repository with the id reference and whatever title you like:
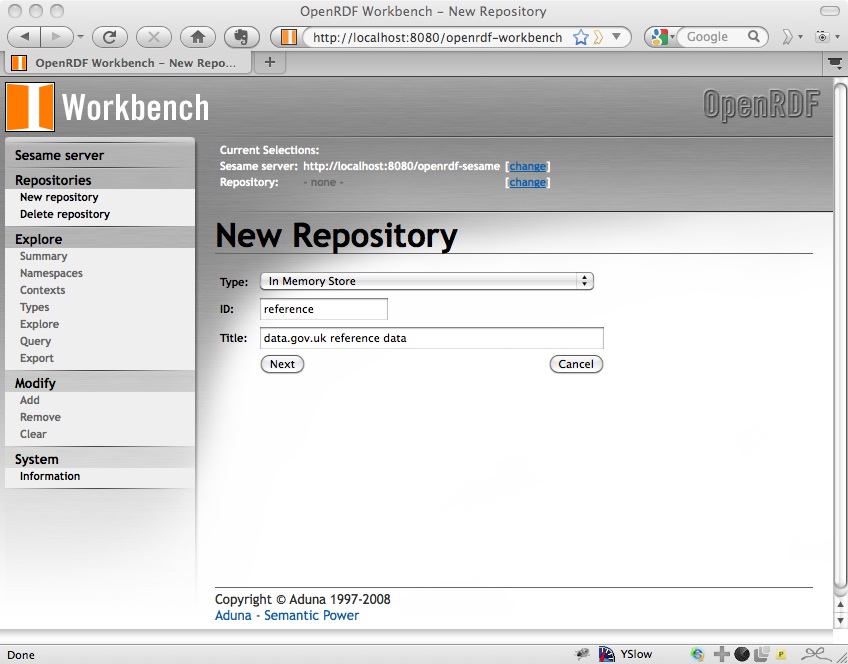
Click Next and there will be a couple more options to select; I just used the default for these:
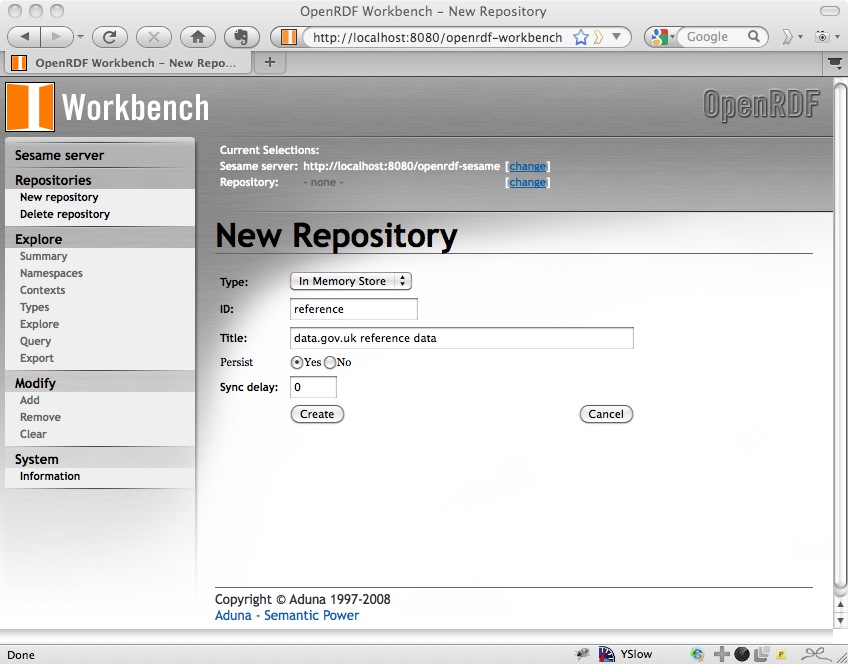
Click Create and you will see a summary of the new repository that you’ve created:
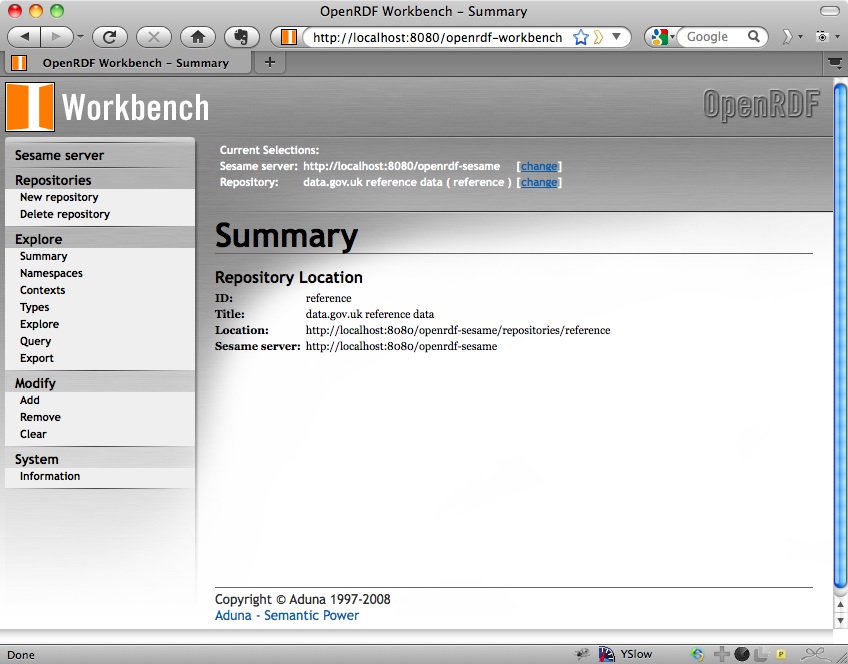
Loading Data
I’m going to use the same data as I did before:
http://source.data.gov.uk/data/reference/organogram-co/2010-10-31/index.rdf
You can add data to a Sesame repository in a browser through the Workbench by uploading a file, pointing Sesame at a URL or pasting in some RDF that you want to load. There are also Java bindings for adding data to Sesame. But neither of those are any good to us as we need programmatic access.
So we will use the HTTP method. I want to add some statements to the reference repository in the graph (what Sesame calls “context”) http://source.data.gov.uk/data/reference/organogram-co/2010-10-30, which amounts to an HTTP PUT on the repository’s statements with that context.
Now I don’t know much at all about Python, but it looks as though the built-in library urllib2 doesn’t support PUT and there’s a better HTTP library available in httplib2. MacPorts supports various different packages for httplib2 with different versions of Python. Now there only seems to be a package for rdflib, which we’ll use later, for Python 2.6, so we’ll go for py26-httplib2, which will bring in Python 2.6 with it just in case.
$ sudo port install py26-httplib2
After running this, if you want to actually use it you will need to install the python_select port and choose Python 2.6:
$ sudo port install python_select
$ sudo python_select python26
Then open up another Terminal window or tab (because the change won’t have affected your old one) and check what version of Python you’re running:
$ python --version
Python 2.6.6
With the httplib2 library in place, it’s time for a Python script (load-rdf-into-sesame.py) to do the PUTting:
import urllib
import httplib2
repository = 'reference'
graph = 'http://source.data.gov.uk/data/reference/organogram-co/2010-06-30'
filename = '/Users/Jeni/Downloads/index.rdf'
print "Loading %s into %s in Sesame" % (filename, graph)
params = { 'context': '<' + graph + '>' }
endpoint = "http://localhost:8080/openrdf-sesame/repositories/%s/statements?%s" % (repository, urllib.urlencode(params))
data = open(filename, 'r').read()
(response, content) = httplib2.Http().request(endpoint, 'PUT', body=data, headers={ 'content-type': 'application/rdf+xml' })
print "Response %s" % response.status
print content
Run the script from the command line:
$ python load-rdf-into-sesame.py
and you should get just get:
Loading /Users/Jeni/Downloads/index.rdf into http://source.data.gov.uk/data/reference/organogram-co/2010-06-30 in Sesame
Response 204
which isn’t particularly helpful (well, the 204 response tells us it worked), but you can then check http://localhost:8080/openrdf-workbench/repositories/reference/contexts and you should see that there is a new context of http://source.data.gov.uk/data/reference/organogram-co/2010-06-30:
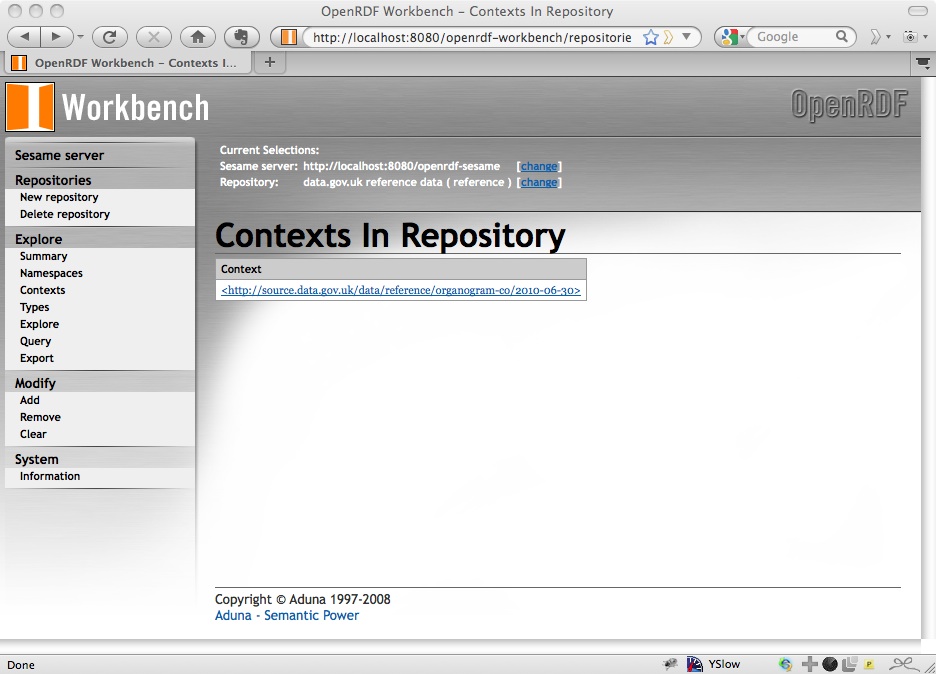
Click on the context and it will take you to a list of (some of) the triples in that graph:
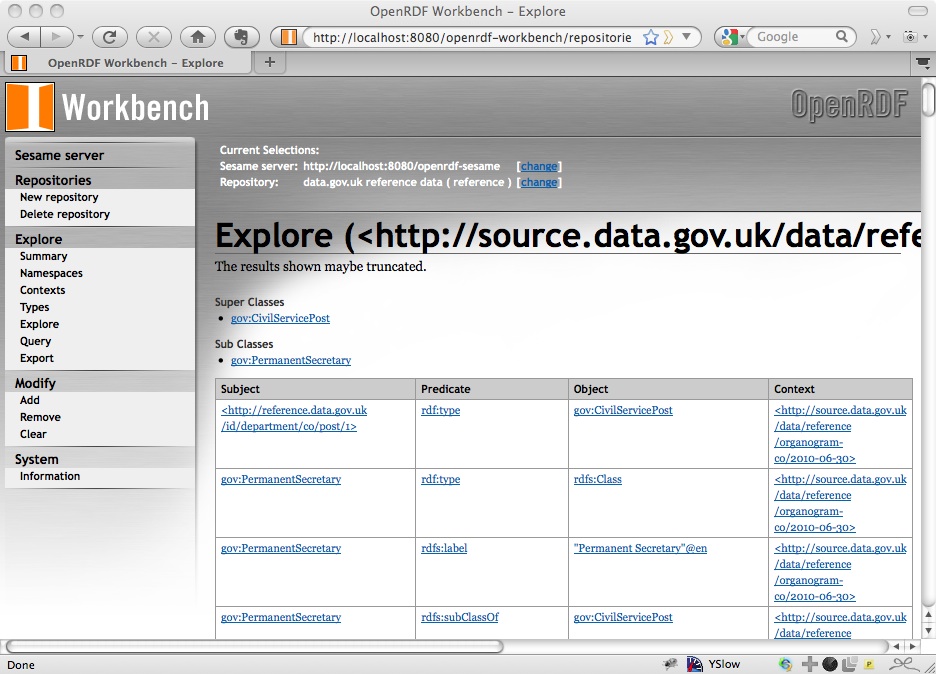
One of the nice things about Sesame is that the Workbench provides you with ways of exploring the data that you have loaded. On the left navigation bar there are ways of listing the types of the entities described in the data:
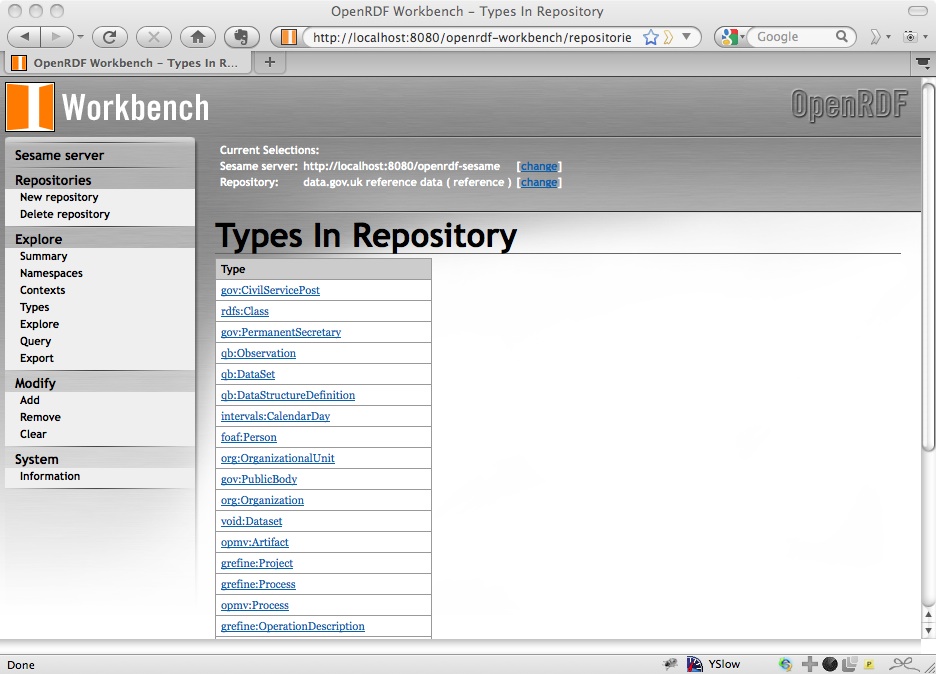
from which you can find instances of that type, for example of org:Organization:
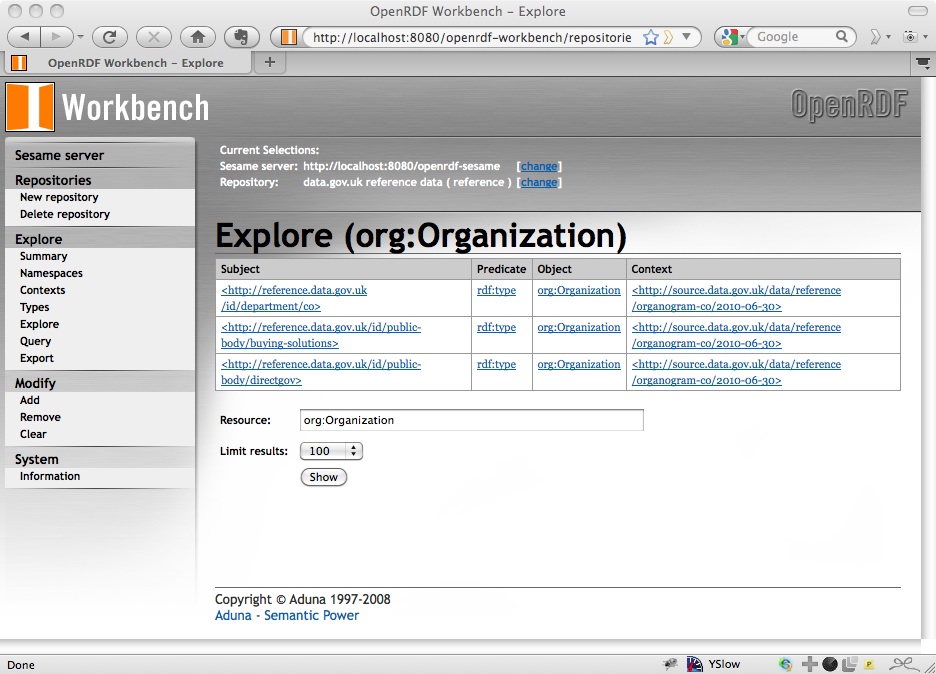
and then the statements about a particular instance, for example DirectGov:
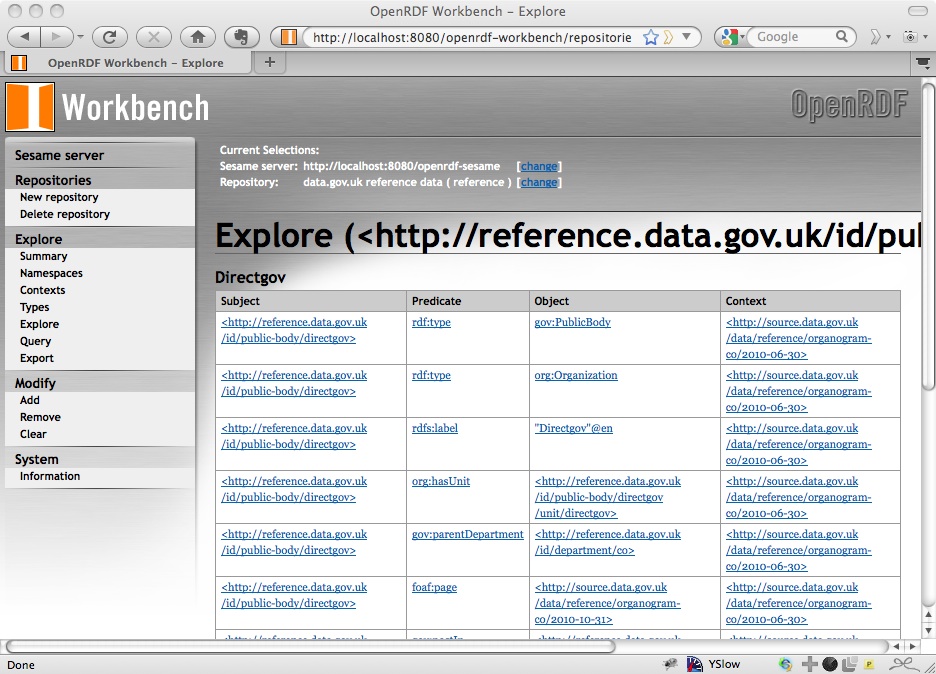
Running a Query
Onto running a query directly. This is done on Sesame in exactly the same way as it was done on 4store in my last walkthrough: by HTTP POSTing a query to the SPARQL endpoint. Sesame’s page for testing queries on the reference repository is at http://localhost:8080/openrdf-workbench/repositories/reference/query and we’ll use the basic one that lists types of things that are described within the data:
SELECT DISTINCT ?type
WHERE {
?thing a ?type .
}
ORDER BY ?type
Paste that into the textarea that’s provided on http://localhost:8080/openrdf-workbench/repositories/reference/query so it looks like:
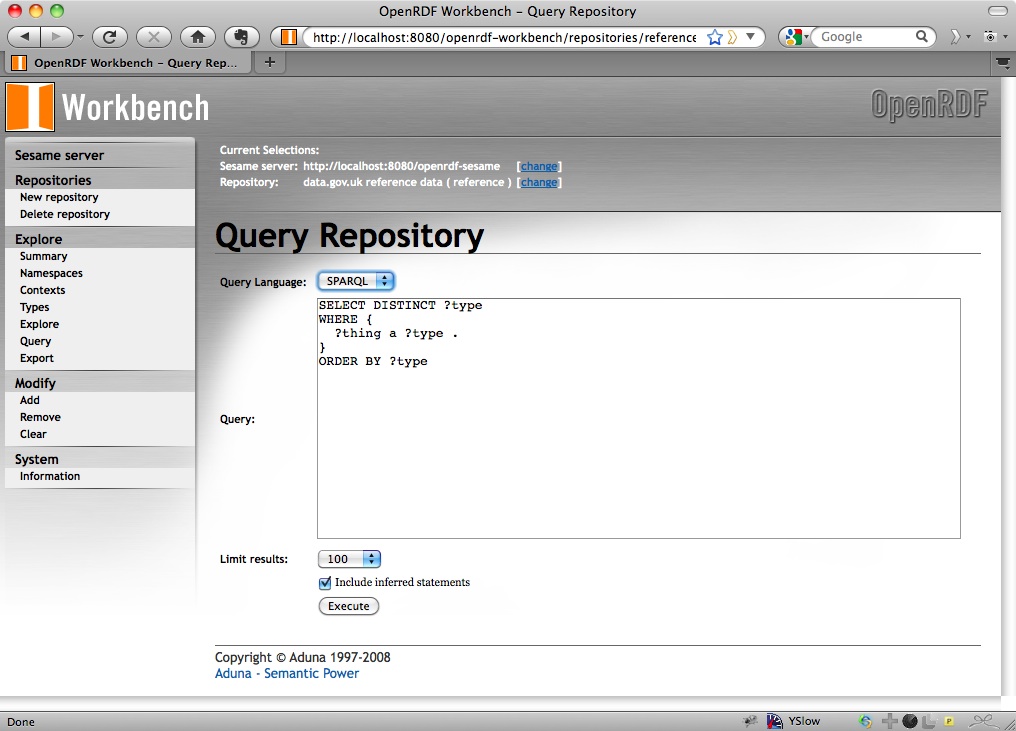
and you get an HTML page:
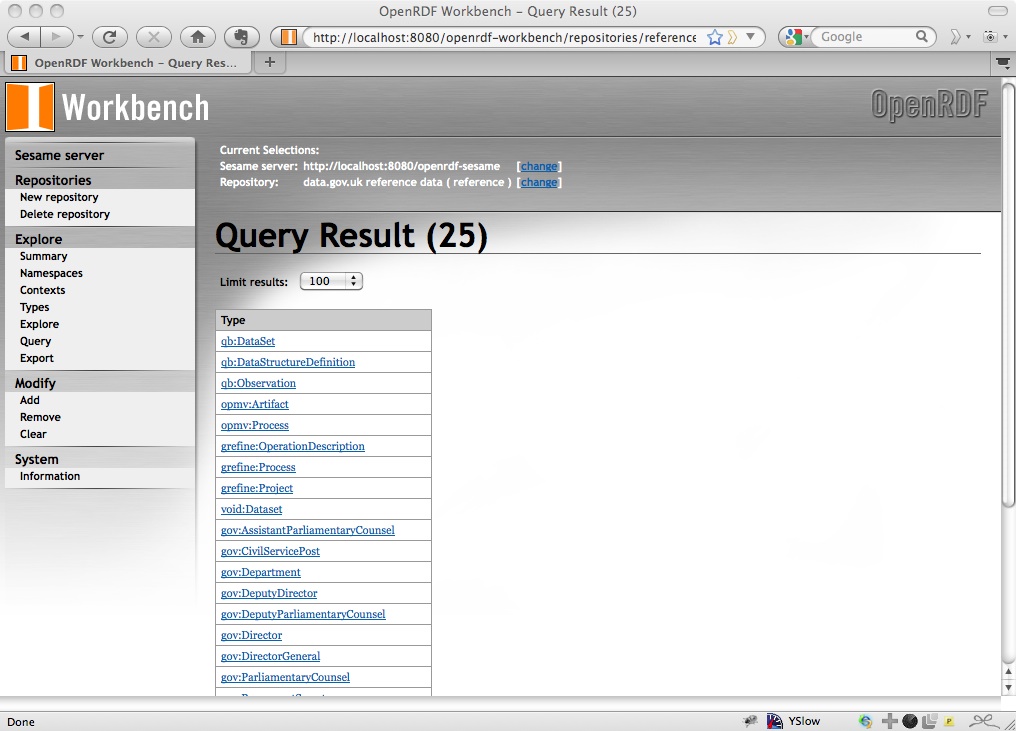
That’s nice for humans, but not so good for computers. When we request the results of this query programmatically, we’ll want to make sure that we specifically ask for the query results in either XML or JSON.
I went the XML route last time, so let’s mix it up a bit and try processing the JSON results of a SPARQL query this time, as it’s really easy to access using the json module in Python. So, we need to POST the query, ensuring that we set the Accept header to application/sparql-results+json, and then process the results as JSON. Here is find-rdf-types.py
import urllib
import httplib2
import json
query = 'SELECT DISTINCT ?type WHERE { ?thing a ?type . } ORDER BY ?type'
repository = 'reference'
endpoint = "http://localhost:8080/openrdf-sesame/repositories/%s" % (repository)
print "POSTing SPARQL query to %s" % (endpoint)
params = { 'query': query }
headers = {
'content-type': 'application/x-www-form-urlencoded',
'accept': 'application/sparql-results+json'
}
(response, content) = httplib2.Http().request(endpoint, 'POST', urllib.urlencode(params), headers=headers)
print "Response %s" % response.status
results = json.loads(content)
print "\n".join([result['type']['value'] for result in results['results']['bindings']])
Run it:
$ python find-rdf-types.py
and you get:
POSTing SPARQL query to http://localhost:8080/openrdf-sesame/repositories/reference
Response 200
http://purl.org/linked-data/cube#DataSet
http://purl.org/linked-data/cube#DataStructureDefinition
http://purl.org/linked-data/cube#Observation
http://purl.org/net/opmv/ns#Artifact
http://purl.org/net/opmv/ns#Process
http://purl.org/net/opmv/types/google-refine#OperationDescription
http://purl.org/net/opmv/types/google-refine#Process
http://purl.org/net/opmv/types/google-refine#Project
http://rdfs.org/ns/void#Dataset
http://reference.data.gov.uk/def/central-government/AssistantParliamentaryCounsel
http://reference.data.gov.uk/def/central-government/CivilServicePost
http://reference.data.gov.uk/def/central-government/Department
http://reference.data.gov.uk/def/central-government/DeputyDirector
http://reference.data.gov.uk/def/central-government/DeputyParliamentaryCounsel
http://reference.data.gov.uk/def/central-government/Director
http://reference.data.gov.uk/def/central-government/DirectorGeneral
http://reference.data.gov.uk/def/central-government/ParliamentaryCounsel
http://reference.data.gov.uk/def/central-government/PermanentSecretary
http://reference.data.gov.uk/def/central-government/PublicBody
http://reference.data.gov.uk/def/central-government/SeniorAssistantParliamentaryCounsel
http://reference.data.gov.uk/def/intervals/CalendarDay
http://www.w3.org/2000/01/rdf-schema#Class
http://www.w3.org/ns/org#Organization
http://www.w3.org/ns/org#OrganizationalUnit
http://xmlns.com/foaf/0.1/Person
This is the same set of types as that given through the HTML browse interface. Note that the JSON results themselves look like:
{
"head": {
"vars": [ "type" ]
},
"results": {
"bindings": [
{
"type": { "type": "uri", "value": "http:\/\/purl.org\/linked-data\/cube#DataSet" }
},
{
"type": { "type": "uri", "value": "http:\/\/purl.org\/linked-data\/cube#DataStructureDefinition" }
},
{
"type": { "type": "uri", "value": "http:\/\/purl.org\/linked-data\/cube#Observation" }
},
...
]
}
}
Each of the items within the bindings array contains a set of bindings for the variables in the SPARQL query. This closely matches the XML format.
Processing RDF
Now we get onto the part of this where we look at specific libraries for RDF support in Python. The most popular library is rdflib, which you can install using MacPorts:
$ sudo port install py26-rdflib
The SPARQL query we’ll try this time uses a CONSTRUCT query, which creates RDF, rather than a SELECT query, which as we’ve seen can create either XML or JSON. For example, try the query:
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
CONSTRUCT {
?person
a foaf:Person ;
foaf:name ?name ;
?prop ?value .
} WHERE {
?person a foaf:Person ;
foaf:name ?name ;
?prop ?value .
}
This gets all the information in the data about the individuals for whom names have been supplied in the data, as RDF. Again, Sesame will display this as HTML when you try doing it, but you can choose a different format from the drop-down menu at the top of the Query Result display:
When you’re not accessing using a browser, by default Sesame serves up its results in TriG format, which isn’t particularly appropriate for the results of CONSTRUCT queries as we don’t need multiple graphs. We’ll request N-Triples as that’s something rdflib can understand. Sesame 2 uses the content type text/plain for N-Triples, so we can request this type by setting the Accept header:
params = { 'query': query }
headers = {
'content-type': 'application/x-www-form-urlencoded',
'accept': 'text/plain'
}
(response, content) = httplib2.Http().request(endpoint, 'POST', urllib.urlencode(params), headers=headers)
We then need to parse this Turtle response into a rdflib.Graph object:
graph = rdflib.ConjunctiveGraph()
graph.parse(rdflib.StringInputSource(content), format="nt")
We then need to get information out of that graph, which rdflib isn’t particularly good at. So let’s use RDFAlchemy instead. That can be installed using easy_install:
$ sudo easy_install-2.6 rdfalchemy
RDFAlchemy can be used to map RDF graphs onto Python object structures in a fairly straight-forward manner. Basically, you define the namespaces of the vocabularies that you want to use, then some classes for the kinds of things that you have in the data, with properties that map onto properties in the RDF. Then you set the rdfSubject.db to the source of the data (which can be an rdflib Graph as above) and can query on it. Here’s an example:
FOAF = rdflib.Namespace('http://xmlns.com/foaf/0.1/')
RDF = rdflib.Namespace('http://www.w3.org/1999/02/22-rdf-syntax-ns#')
class Person(rdfalchemy.rdfSubject):
rdf_type = FOAF.Person
name = rdfalchemy.rdfSingle(FOAF.name)
mbox = rdfalchemy.rdfSingle(FOAF.mbox)
rdfalchemy.rdfSubject.db = graph
stott = Person.get_by(name='Andrew Stott')
print "Andrew Stott's email address: %s" % stott.mbox.n3()
RDFAlchemy adds both get_by() and filter_by() methods on the descriptor classes that you define, to get a single item that matches a query or a list of items, respectively.
The full script for ‘get-names-and-emails.py’ is:
import urllib
import httplib2
import rdflib
import rdfalchemy
query = """PREFIX foaf: <http://xmlns.com/foaf/0.1/>
CONSTRUCT {
?person
a foaf:Person ;
foaf:name ?name ;
?prop ?value .
} WHERE {
?person a foaf:Person ;
foaf:name ?name ;
?prop ?value .
}"""
repository = 'reference'
endpoint = "http://localhost:8080/openrdf-sesame/repositories/%s" % repository
print "POSTing SPARQL query to %s" % endpoint
params = { 'query': query }
headers = {
'content-type': 'application/x-www-form-urlencoded',
'accept': 'text/plain'
}
(response, content) = httplib2.Http().request(endpoint, 'POST', urllib.urlencode(params), headers=headers)
print "Response %s" % response.status
graph = rdflib.ConjunctiveGraph()
graph.parse(rdflib.StringInputSource(content), format="nt")
print "Loaded %d triples" % len(graph)
FOAF = rdflib.Namespace('http://xmlns.com/foaf/0.1/')
RDF = rdflib.Namespace('http://www.w3.org/1999/02/22-rdf-syntax-ns#')
class Person(rdfalchemy.rdfSubject):
rdf_type = FOAF.Person
name = rdfalchemy.rdfSingle(FOAF.name)
mbox = rdfalchemy.rdfSingle(FOAF.mbox)
rdfalchemy.rdfSubject.db = graph
stott = Person.get_by(name='Andrew Stott')
print "Andrew Stott's email address: %s" % stott.mbox.n3()
Run this script with:
$ python get-names-and-emails.py
and you get the result:
No handlers could be found for logger "rdflib.Literal"
POSTing SPARQL query to http://localhost:8080/openrdf-sesame/repositories/reference
Response 200
Loaded 459 triples
Andrew Stott's email address: <mailto:andrew.stott@cabinet-office.gsi.gov.uk>
The first line is apparently a side-effect of rdflib/RDFAlchemy weirdness which you don’t need to worry about. The rest shows that the search was successful; the call to the .n3() call on the email address is only necessary because it is a resource rather than a literal, and therefore doesn’t get converted to a particularly readable string otherwise.
Conclusions
So there you have it, another walkthrough of setting up a local triplestore, loading in data and accessing that data programmatically using SPARQL queries, this time using Sesame and Python rather than 4store and Ruby.
This walkthrough took me a fair bit longer to do than the previous one, for several reasons:
- I’ve done almost no previous programming with Python (as you can probably tell), so every little thing took ages to work out – you know you’re in trouble when you’re Googling for string concatenation code! I’m very happy to accept corrections and improvements, which I’ll include in the above.
- I spent a lot of time faffing around with different Python versions, opting for the latest and then finding that the libraries that I wanted to use weren’t available for that version and so on. I eventually ended up with Python 2.6; the code above may or may not work with any other versions.
- Setting up Sesame 2 was pretty frustrating: first Tomcat wouldn’t work, then Jetty wouldn’t work, and finally I did get Tomcat working and then had the issue with the log directory, as I described above. Once I’d changed the data directory things worked very smoothly.
- I thought rdflib was going to be enough to work with RDF in Python, but really it isn’t (if you want to get data out as well as put data in), so I had to find something else.
- The documentation for rdflib and RDFAlchemy isn’t as comprehensive as the documentation for RDF.rb, especially if you’re not familiar with Python, so it took me a bit longer to work out how to do things with those particular libraries.
- I took a lot more screenshots!
Again, I haven’t followed Richard’s steps to the letter; in particular I haven’t used a package to get data out of (or into) Sesame: I’ve just done it through HTTP calls. I did it this way deliberately because I think it’s a really important feature of triplestores that you can query them through a common interface: SPARQL. It means that you can take the Python code here and use it against 4store or another triplestore with only a change to the value of the endpoint variable, and similarly take the Ruby code from my previous walkthrough and use it against Sesame. Your code is not tied to a particular implementation or API; you “only” have to learn SPARQL and you’re away.
If you prefer something a little more tightly bound, however, RDFAlchemy does have some targeted Sesame support, as does RDF.rb for that matter. These can help with the management of the data within the repository as well as querying it.
Another thing that’s worth pointing out is that 4store and Sesame have completely different (HTTP-based) interfaces for getting data into stores, and that rdflib/RDFAlchemy and RDF.rb have completely different ways of loading data into in-memory graphs, querying it, and getting information from the results, quite aside from the obvious language-based differences that you’d expect.
On the SPARQL side, there are some efforts within the W3C to define a uniform HTTP protocol for managing RDF graphs and of course there’s SPARQL 1.1 Update. There are glimmers of hope for a standard RDF API, as I’ve argued for recently, but I gather that this effort will be focused on client-side developers, ie that it is really a standard RDF API for Javascript, which I think is a wasted opportunity: I would have been faster in this task if I’d been able to use familiar methods, and I wouldn’t have been so dependent on the documentation provided by the author of a particular library.
Anyway, hopefully my tramping this path will make it easier for those who follow.